Abstract
Background: Clinical prediction rules (CPRs) can assist clinicians by focusing their clinical evaluation on the most important signs and symptoms, and if used properly can reduce the need for diagnostic testing. This study aims to perform an updated systematic review of clinical prediction rules and classification and regression tree (CART) models for the diagnosis of influenza.
Methods: We searched PubMed, CINAHL, and EMBASE databases. We identified prospective studies of patients presenting with suspected influenza or respiratory infection and that reported a CPR in the form of a risk score or CART-based algorithm. Studies had to report at a minimum the percentage of patients in each risk group with influenza. Studies were evaluated for inclusion and data were extracted by reviewers working in parallel. Accuracy was summarized descriptively; where not reported by the authors the area under the receiver operating characteristic curve (AUROCC), predictive values, and likelihood ratios were calculated.
Results: We identified 10 studies that presented 14 CPRs. The most commonly included predictor variables were cough, fever, chills and/or sweats, myalgias, and acute onset, all which can be ascertained by phone or telehealth visit. Most CPRs had an AUROCC between 0.7 and 0.8, indicating good discrimination. However, only 1 rule has undergone prospective external validation, with limited success. Data reporting by the original studies was in some cases inadequate to determine measures of accuracy.
Conclusions: Well-designed validation studies, studies of interrater reliability between telehealth an in-person assessment, and studies using novel data mining and artificial intelligence strategies are needed to improve diagnosis of this common and important infection.
- Clinical Decision Rules
- Clinical Medicine
- Influenza
- Physical Examination
- Prospective Studies
- Respiratory Diseases
- Systematic Reviews
Introduction
Seasonal influenza accounts for losses in workforce productivity, a strain on health services, and an average of approximately 40,000 deaths1 and a range of 140,000 to 710,000 hospitalizations2 in the United States every year. Neuraminidase inhibitors such as zanamivir and oseltamivir modestly reduce the duration of symptoms by about 30 hours if initiated within 24 hours, and by 20 hours if initiated within 36 hours of symptoms onset for both influenza virus type A and B.3,4 Therefore, if prescribed prompt diagnosis of influenza is needed for maximal benefit and to institute infection control measures such as mask wearing and social distancing.5
Clinical prediction rules (CPRs) use a combination of signs, symptoms, and sometimes simple point-of-care tests to assist diagnostic and therapeutic decisions. They can take the form of a risk score (point score) that is typically derived from the β coefficients of a logistic regression, or as a nonparametric classification and regression tree (CART) model that has as its output a decision tree. As has been demonstrated with other conditions such as sore throat,6 pulmonary embolism,7,8,9 and ankle injury,10 clinical prediction rules can help establish a patient's probability of a given condition and inform the interpretation of subsequent diagnostic tests. They have the potential to reduce the need for diagnostic tests, as patients initially classified as low probability for the condition in question may not require further testing or evaluation, and patients with high probability for the condition may be directed to proceed with therapy or other intervention.
Systematic reviews have been performed of individual signs and symptoms of influenza, but were limited by the search strategy, by not using modern analytic methods for diagnostic meta-analysis, and by their age.11,12 A systematic review of CPRs for the diagnosis of influenza found that the positive predictive value of simple clinical heuristics such as “cough and fever” (26% to 87%) and “cough, fever, and acute onset” (30% to 77%) varied widely between studies.13 However, that systematic review had similar limitations to the meta-analyses of individual signs and symptoms. It also failed to identify more sophisticated risk scores or algorithms. Our goal is therefore to perform an updated, comprehensive systematic review and meta-analysis of clinical prediction rules for the diagnosis of influenza.
Methods
This study is a systematic review of previously published studies of the accuracy of CPRs for the diagnosis of influenza. A CPR was defined as a point score or algorithm (such as those that result from a classification and regression tree analysis) for the diagnosis of laboratory-confirmed influenza. The study was registered with the PROSPERO database (#CRD42020161801) and followed PRISMA guidance regarding conduct and reporting of a diagnostic meta-analysis.
Data Sources and Searches
We searched PubMed, CINAHL and EMBASE databases in August 2020, with a bridge search in June 2021. The primary search strategy is shown in Appendix 1. It included individual signs and symptoms, whose accuracy is reported in a separate systematic review (manuscript in preparation). We also used a recently developed search strategy designed to be highly sensitive for detection of CPRs, shown in Appendix 2.14 The limits “has abstract” and “human” were applied to both searches. In addition, the reference lists of included studies were reviewed for additional articles, as were 3 older systematic reviews identified by our search.10⇓–12
Study Selection
Studies were included if they recruited a prospective cohort of children, adolescents, or adults presenting with symptoms of respiratory tract infection (RTI) or clinically suspected influenza in the inpatient or outpatient setting. Studies had to report sufficient information to calculate the accuracy of a risk score or classification tree for the diagnosis of laboratory-confirmed influenza; studies presenting a multivariate model not easily used by clinicians were excluded. Simple heuristics that studied the presence or absence of 2 or 3 symptoms for the diagnosis of influenza, such as “cough and fever indicates influenza,” were excluded from this study but are addressed in a companion systematic review (manuscript in preparation). No limits were set for country or year; language was limited to publications in English. The reference standard had to be polymerase chain reaction (PCR), culture or serology and had to have been performed in all participants to avoid verification bias. Studies also had to report at a minimum the percentage of patients in each risk group with influenza.
Studies were excluded if they did not enroll patients with acute RTI or suspected influenza, or if they were in a specialized population such as only patients in skilled nursing facilities, immunosuppressed patients, or patients with chronic lung disease. Studies were also excluded if they used a case-control design (ie, recruited patients with known influenza and healthy controls) as this has been shown to inflate the apparent accuracy of diagnostic tests.15 Finally, studies that reported a multivariate model but did not convert it to a point score that is easily usable by clinicians were excluded.
Data Extraction and Quality Assessment
All abstracts were reviewed for inclusion in parallel by 2 authors, 1 of whom was a physician. For any abstract deemed potentially of interest by any reviewer, the full article was obtained and reviewed by the corresponding author and 1 other reviewer. Studies meeting inclusion and exclusion criteria were reviewed in parallel by 2 authors who each abstracted study characteristics including the type of validation (if any), study quality, and test accuracy data (true positive, false positive, false negative, and true positive in comparison with a valid reference standard). Discrepancies were resolved through consensus discussion. The Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool was adapted for our study and definitions for low, unclear and high risk of bias prespecified for each domain as shown in Appendix 3.14
Data Synthesis and Analysis
For published CPRs, the probability of influenza in each risk group, likelihood ratios, and the proportion of patients in that risk group are reported. If a CPR reported more than 2 risk groups, the likelihood ratio was calculated for each risk group (“stratum specific likelihood ratios”). If a point score was proposed without identification of low, moderate and high-risk groups, we used clinical judgment to propose such groups post hoc based on test and treatment thresholds from a previous study and reported those findings separately from the original study findings. For published studies of a point score but reporting the area under the receiver operating characteristic curve (AUROCC), AUROCC was calculated using the pROC package in R.16
Results
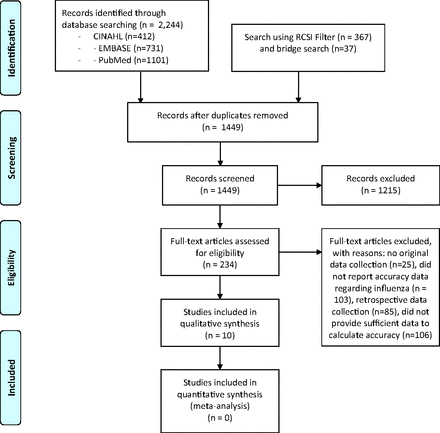
The search strategy described in Appendix 1 yielded 1209 unique studies from 3 databases, and the initial and bridge searches using the RCSI filter for clinical prediction rule searches in Appendix 2 identified 404 studies, of which 240 were unique for a total of 1449 unique studies. After screening abstracts, the full text of 234 studies was reviewed to determine whether they met the inclusion and exclusion criteria for the systematic review. A total of 10 met those criteria and reported the accuracy of a CPR (either a point score or a classification tree) for influenza. Our review of previous systematic reviews identified no additional studies. Therefore, a total of 10 studies met our inclusion criteria and were included in the analysis. See Figure 1 for the PRISMA flow diagram.
PRISMA diagram describing the search.
Study Characteristics
Characteristics of the included studies are summarized in Table 1. Published stiudies ranged in size from 456 to 4572 patients, and all were based on outpatient populations. Four studies were set in North America, 2 in Europe, 2 in Asia, and 2 publications used a combined dataset from Switzerland and the United States. The reference standard test was PCR for 7 studies, serology for 1, and culture or PCR for 2. Most tested for both influenza A and B and the prevalence of laboratory-confirmed influenza ranged from 6.8% to 34.2%.
Characteristics of Included Studies That Develop or Validate Clinical Prediction Rules for Influenza
These 10 studies reported a total of 7 CART algorithms and 7 risk scores, summarized in Appendix 4. Two studies used the same dataset.17,18 For 2 studies, we proposed a new “post hoc” risk score for each 1 using their data based on our assessment of what we considered to be clinically useful low, moderate and high-risk groups.19,20
Study Quality
Assessment of study quality is summarized in Table 2. Six studies were judged to be at low risk of bias, while 4 were judged at moderate risk of bias. For the latter, in 1 dataset (reported by 2 studies) the potential bias was because 2 different reference standards were used (culture and PCR).17,18 A second study did not account for all patients at the end of the study,21 while a third study did not use PCR for any patients.19
Evaluation of Study Quality Using the QUADAS-2 Framework for Studies of Influenza Clinical Prediction Rules
Validation Procedures
With regards to validation of proposed CPRs, 2 studies performed no validation and only presented data from the derivation population.20,22 Five studies performed internal validation using a split sample;17,18,21,23,24 this approach randomly divides a single dataset into 2 groups, 1 to derive the CPR and 1 to validate or test it. This approach is considered less robust than true external validation in a completely new population, since the characteristics of the patients in the 2 groups are very similar.25 One study performed prospective temporal internal validation, developing the score with data from 2009 to 2013 and validating it with data from the same clinical center in 2014.26 The preferred approach of external, prospective validation in a completely new population was performed for a single risk score, the FluScore, in 1 published study.27
Predictor Variables
The predictor variables in each risk score or CART model are summarized in Table 3. Cough, fever, chills and/or sweats, and myalgias, were present in between 7 and 12 models. Acute onset, coryza, sore throat, age, headache, and fatigue or malaise were present in between 2 and 4 models, while sneezing, whether influenza was circulating in the community, close contact with influenza, male sex, and sinus problems were each present in a single model.
Predictor Variables Included in Each Influenza Risk Score or Classification and Regression Tree (CART) Model
Accuracy of Clinical Prediction Rules
The accuracy of CPRs varied widely; it is summarized in Table 4. The AUROCC ranged from 0.64 to 0.79 in studies of risk scores, and was not reported by 4 studies. The AUROCC ranged from 0.68 to 0.82 for CART models. The proportion of patients classified as low risk by a CPR ranged from 16% to 89%, with a wide range of probabilities of influenza in these low-risk groups depending on the risk score and the overall prevalence of influenza in the population.
Accuracy of Clinical Prediction Rules for Influenza
Validation of Clinical Prediction Rules
Two CPRs were prospectively validated. Van Vugt and colleagues used data from the European multi-center GRACE study to validate the FluScore.27 The AUROCC was 0.79 in the original study (combining both derivation and validation) and 0.71 in the prospective validation. The stratum specific likelihood ratios were 0.10/1.02/2.47 and 0.51/2.53/3.24 for the original and prospective validation groups respectively. More patients were classified as low risk in the study by van Vugt and colleagues, 27 which is clinically useful for telephone or other triage, but this group had a 13.6% prevalence of influenza compared with 5.0% in the original validation.
Anderson and colleagues developed a CART model using data from 3782 patients identified in 2009 to 2013, and validated it in the same population using 2014 data for 790 patients.26 They report only an overall AUROCC of 0.69 for derivation and validation groups combined, and positive likelihood ratio (LR+) of 2.73 and negative likelilhood ratio (LR-) of 0.52 for the derivation group (data to calculate likelihood ratios not reported for the validation group). The prevalence of influenza in their low- and high-risk groups was consistent, 21.9% vs 20.2% and 59.7% vs 63.8% in derivation and validation groups respectively.
Discussion
Our systematic review identified 7 published risk scores and 7 published CART algorithms for diagnosis of influenza. Only 2 of the CPRs have had any kind of prospective validation. One CPR did not validate particularly well,27 but the validation study did not directly ask patients about the presence of chills or sweats, and apparently assumed that they were present in all patients with fever. This assumption could adversely affect validation. 27 The other prospective validation study was prospective temporally but studied patients at the same clinical site,24 which is considered inferior to prospective validation in an entirely new population.25 The overall accuracy of most rules was moderately good based on areas under the ROC curve that were typically between 0.7 and 0.8. Despite being a common condition, the development and especially validation of CPRs for influenza remains poorly studied.25 While the use of anti-influenza drugs is not universally endorsed, making an accurate, early diagnosis could help implement mask wearing and distancing to reduce spread of the disease in the community, as was seen during the COVID-19 pandemic.5
Only 4 CPRs divided patients into low, moderate and high-risk groups,17,18 with the remainder generally identifying only low- and high-risk groups. We would argue that it is most useful to clinicians to identify 3 risk groups, which can potentially correspond to the the 3 groups identified by Pauker and Kassirer's threshold model of diagnosis: below the test threshold (rule out), between the test and treatment thresholds (gather more information), and above the treatment threshold (treat empirically).28 These thresholds have been studied, with 1 study finding a test threshold of 5% for US physicians and 32% for Swiss physicians, and treatment thresholds of 55% and 68% respectively.29 A second study in older adults estimated test and treament thresholds of 10% and 41% respectively.30 The test threshold in particular will be influenced by the availability and adoption of diagnostic testing versus relying on clinical diagnosis alone. A CPR in the US would be most useful if the low-risk group had a very low risk of influenza, so testing could be avoided, while the high-risk group had a probability of influenza of at least 55% so treatment could be started empirically. This is especially relevant for telehealth settings, and when advising patients whether they should be evaluated.
An often overlooked metric for CPRs is the proportion of patients classified as low or high risk. If the CPR is designed properly with test and treatment thresholds in mind, the probability of disease in the low-risk group is low enough that the disease can be ruled out without testing, and the probability of disease in the high-risk group is high enough that empiric therapy may be appropriate. In this case, it is most useful if a larger proportion of patients fall into those low or high-risk groups. The moderate risk group requires further evaluation and potentially testing (for example using a point-of-care influenza test) which might require an in-person visit and is therefore more expensive and burdensome. How low a risk is “low enough” will vary with the test threshold, which as we have previously demonstrated can vary considerably between countries.29 It also likely varies at the individual level, as providers may vary in their perception of risk of influenza complications and/or benefits of treatment or other measures such as quarantine.
The most commonly used predictor variables were cough, fever, chills or sweats, myalgias, acute onset, coryza, sore throat, headache and fatigue. Future studies of influenza diagnosis should consider this a minimum dataset, but should also consider combinations of symptoms as interaction terms in multivariate models, differential weighting by symptom severity, and novel symptoms such as the time to maximum symptoms severity. Different models are likely needed for different age groups, and vaccine status should be included more often in models.31,32
Limitations of our meta-analysis include the failure of many studies to fully report diagnostic accuracy, for example the area under the receiver operating characteristic curve. We were also unable to perform meta-analysis, since with only 1 exception17,27 each study reported a different proposed CPR.
In the COVID-19 pandemic, more and more care is being provided via telehealth.33 This is likely to persist for some time, and if high quality care of respiratory infections such as COVID-19 or influenza can be delivered remotely, there is the potential for telehealth to reduce transmission to others in the outpatient setting. The most commonly used predictor variables (Table 3) can all be ascertained remotely, and could potentially be supplemented by home testing for influenza.34
Future studies should follow reporting recommendations of the “Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis” (TRIPOD) statement including clear definitions of predictors and outcomes, full reporting of models, and prospective validation in new populations.25 In addition, there is a need for prospective validation of existing rules, as well as the development and prospective validation of more CPRs developed using novel machine learning strategies.31,32 Studies of interrater reliability of patient and physician assessment of symptoms of influenza are also needed. In addition to prospectively gathering new data, existing datasets could be pooled for individual patient meta-analysis of diagnosis as well as to develop and internally validate new CPRs. Rules designed to use symptoms only to rule out influenza could be especially useful for telehealth or triage, reducing the need for in-person visits.
Appendix 1. Search for Diagnosis of Influenza Using the History and Physical Examination
Databases Used
PubMed
CINAHL
EMBASE
Keyword Development
We developed keyword combinations to retrieve articles from the databases that contain 4 following components:
Influenza.
Methods related to clinical decision rule/predictive modelling to diagnose Influenza
Laboratory tests as the reference standards to diagnose Influenza.
Sign and symptoms related to Influenza.
Summary of Search Activities
We generated generic terms and subject headings that are specific to each database based on the 4 components of keyword combinations. We applied the combinations of the generic terms and the subject headings differently in each database using their specific algorithm. The date of our last search was on October 14, 2019.
PubMed
- Keyword component #1:
((influenza OR flu [tiab]) OR “Influenza, Human"[Mesh])
- Keyword component #2:
(“predictive"[tiab] OR “predictor*"[tiab] OR “prediction*"[tiab] OR “clinical model"[tiab] OR “clinical score"[tiab] OR “clinical scoring"[tiab] OR “clinical rule"[tiab] OR “decision guideline"[tiab] OR “decision model"[tiab] OR “validation study"[tiab] OR “validation studies"[tiab] OR “derivation study"[tiab] OR “screening score"[tiab] OR “decision rule"[tiab] OR “diagnostic rule"[tiab] OR “diagnostic score"[tiab] OR “predictive outcome"[tiab] OR “predictive rule"[tiab] OR “scoring"[tiab] OR “risk score"[tiab] OR “risk scoring"[tiab] OR “prognostic score"[tiab] OR “prognostic index"[tiab] OR “prognostic rule"[tiab] OR “prospective validation"[tiab] OR “diagnostic model"[tiab] OR “clinical definition"[tiab] OR “case definition*"[tiab] OR “Bayes Theorem"[Mesh] OR “Decision Support Techniques"[Mesh] OR “sensitivity and specificity” [MeSH Terms] OR “predictive value of tests” [MeSH Terms] OR (“risk”[tiab] AND “tool”[tiab]) OR ((“validate”[tiab] OR “validation”[tiab] OR “validating”[tiab] OR “develop”[tiab] OR “development”[tiab] OR “derivation”[tiab] OR “derive”[tiab] OR “deriving”[tiab] OR “performance”[tiab]) AND (“decision”[tiab] OR “predictive”[tiab] OR “prediction”[tiab] OR “rule”[tiab] OR “score"[tiab] OR “scoring”[tiab] OR “index”[tiab] OR “model”[tiab] OR “scale”[tiab] OR “tool”[tiab] OR “algorithm”[tiab])) OR (“development”[tiab] AND “validation”[tiab]) OR (“derivation”[tiab] AND “validation”[tiab]))
- Keyword component #3:
(“gold standard”[tiab] OR “diagnostic testing”[tiab] OR “laboratory”[tiab] OR culture[tiab] OR “virus positive”[tiab] OR immunofluorescence[tiab] OR “viral culture”[tiab] OR “Polymerase Chain Reaction"[Mesh] OR “Clinical Laboratory Techniques"[Mesh] OR “Reverse Transcriptase Polymerase Chain Reaction"[MeSH Terms] OR “Influenza A virus/isolation and purification"[MeSH Terms] OR “Predictive Value of Tests"[MeSH Terms] OR PCR[All Fields])
- Keyword component #4:
(symptom*[tiab] OR sign*[tiab] OR temperature[tiab] OR stiffness[tiab] OR myalgia[tiab] OR rhinorrhea[tiab] OR cough[tiab] OR fever[tiab] OR “sore throat”[tiab] OR headache[tiab] OR “Fever"[Mesh] OR “Cough"[Mesh] OR “Pharyngitis"[Mesh] OR “Headache"[Mesh] OR"Myalgia"[Mesh])
- Filter: English as language and the presence of abstract
- Combined keywords
Search: #1 AND #2 AND #3 AND #4
Filters: Abstract, English
- Results: 1101
CINAHL
- Keyword component (#1):
(MH “Influenza+”) OR TI ((Influenza OR flu)) OR AB ((Influenza OR flu))
- Keyword component (#2):
(MH “Predictive Validity”) OR (MH “Predictive Value of Tests”) OR (MH “Predictive Research”) OR (MH “Cox Proportional Hazards Model”) OR (MH “Multiple Logistic Regression”) OR (MH “Loglinear Models”) OR (MH “Decision Trees”) OR (MH “Validation Studies”) OR TI (“predictive” OR “predictor*” OR “prediction*” OR “clinical model” OR “clinical score” OR “clinical scoring” OR “clinical rule” OR “decision guideline” OR “decision model” OR “validation study” OR “validation studies” OR “derivation study” OR “screening score” OR “decision rule” OR “diagnostic rule” OR “diagnostic score” OR “predictive outcome” OR “predictive rule” OR “scoring” OR “risk score” OR “risk scoring” OR “prognostic score” OR “prognostic index” OR “prognostic rule” OR “prospective validation” OR “diagnostic model” OR “clinical definition” OR “case definition*”) OR AB (“predictive” OR “predictor*” OR “prediction*” OR “clinical model” OR “clinical score” OR “clinical scoring” OR “clinical rule” OR “decision guideline” OR “decision model” OR “validation study” OR “validation studies” OR “derivation study” OR “screening score” OR “decision rule” OR “diagnostic rule” OR “diagnostic score” OR “predictive outcome” OR “predictive rule” OR “scoring” OR “risk score” OR “risk scoring” OR “prognostic score” OR “prognostic index” OR “prognostic rule” OR “prospective validation” OR “diagnostic model” OR “clinical definition” OR “case definition*”) OR TI ((“risk” AND “tool”)) OR AB ((“risk” AND “tool”)) OR TI (((“validate” OR “validation” OR “validating” OR “develop” OR “development” OR “derivation” OR “derive” OR “deriving” OR “performance”) AND (“decision” OR “predictive” OR “prediction” OR “rule” OR “score” OR “scoring” OR “index” OR “model” OR “scale” OR “tool” OR “algorithm”))) OR AB (((“validate” OR “validation” OR “validating” OR “develop” OR “development” OR “derivation” OR “derive” OR “deriving” OR “performance”) AND (“decision” OR “predictive” OR “prediction” OR “rule” OR “score” OR “scoring” OR “index” OR “model” OR “scale” OR “tool” OR “algorithm”))) OR TI ((“development” AND “validation”) OR (“derivation” AND “validation”)) OR AB ((“development” AND “validation”) OR (“derivation” AND “validation”))
- Keyword component (#3):
(MH “Signs and Symptoms+”) OR (MH “Body Temperature Changes+”) OR (MH “Fever+”) OR (MH “Cough”) OR (MH “Muscle Pain”) OR (MH “Pharyngitis”) OR (MH “Headache+”) OR TI (symptom OR symptoms OR sign OR signs OR temperature OR stiffness OR myalgia OR rhinorrhea OR cough OR fever OR “sore throat” OR headache) OR AB (symptom OR symptoms OR sign OR signs OR temperature OR stiffness OR myalgia OR rhinorrhea OR cough OR fever OR “sore throat” OR headache)
- Keyword component (#4):
(MH “Diagnosis, Laboratory+”) OR (MH “Polymerase Chain Reaction+”) OR (MH “Reverse Transcriptase Polymerase Chain Reaction”) OR (MH “Microbial Culture and Sensitivity Tests”) OR (MH “Cell Culture Techniques”) OR (MH “Fluorescent Antibody Technique+”) OR TI (“gold standard” OR “diagnostic testing” OR “laboratory” OR culture OR “virus positive” OR immunofluorescence OR “viral culture” OR PCR) OR AB (“gold standard” OR “diagnostic testing” OR “laboratory” OR culture OR “virus positive” OR immunofluorescence OR “viral culture” OR PCR)
- Filter: None
- Combined keywords:
#1 AND #2 AND #3 AND #4
- Results: 412
EMBASE
- Keyword component (#1):
influenza:ab,ti OR ‘influenza’/exp
- Keyword component (#2):
‘prediction’/exp OR ‘predictive value’/exp OR ‘predictor variable’/exp OR ‘predictive validity’/exp OR ‘bayes theorem’/exp OR ‘sensitivity and specificity’/exp OR ‘clinical model’:ab,ti OR ‘clinical score’:ab,ti OR ‘clinical scoring’:ab,ti OR ‘clinical rule’:ab,ti OR ‘decision guideline’:ab,ti OR ‘decision model’:ab,ti OR ‘validation study’:ab,ti OR ‘validation studies’:ab,ti OR ‘derivation study’:ab,ti OR ‘screening score’:ab,ti OR ‘decision rule’:ab,ti OR ‘diagnostic rule’:ab,ti OR ‘diagnostic score’:ab,ti OR ‘predictive outcome’:ab,ti OR ‘predictive rule’:ab,ti OR ‘scoring’:ab,ti OR ‘risk score’:ab,ti OR ‘risk scoring’:ab,ti OR ‘prognostic score’:ab,ti OR ‘prognostic index’:ab,ti OR ‘prognostic rule’:ab,ti OR ‘prospective validation’:ab,ti OR ‘diagnostic model’:ab,ti OR ‘clinical definition’:ab,ti OR ‘case definition*’:ab,ti
- Keyword component (#3):
‘gold standard’/exp OR ‘dyes, reagents, indicators, markers and buffers’/exp OR ‘polymerase chain reaction’/exp OR ‘reverse transcription polymerase chain reaction’/exp OR ‘immunofluorescence’/exp OR ‘virus culture’/exp OR ‘gold standard’:ab,ti OR ‘diagnostic testing’:ab,ti OR ‘laboratory’:ab,ti OR culture:ab,ti OR ‘virus positive’:ab,ti OR immunofluorescence:ab,ti OR ‘viral culture’:ab,ti
- Keyword component (#4):
‘fever’/exp OR ‘coughing’/exp OR ‘pharyngitis’/exp OR ‘headache’/exp OR ‘myalgia’/exp OR ‘rhinorrhea’/exp OR ‘physical disease by body function’/exp OR symptom:ab,ti OR symptoms:ab,ti OR sign:ab,ti OR signs:ab,ti OR temperature:ab,ti OR stiffness:ab,ti OR myalgia:ab,ti OR rhinorrhea:ab,ti OR cough:ab,ti OR fever:ab,ti OR ‘sore throat’:ab,ti OR headache:ab,ti
- Filter: None
- Combined keywords:
#1 AND #2 AND #3 AND #4
- Results: 731
Appendix 2. Search for clinical prediction rules using RCSI filter1
"Influenza"[ti] and
(“diagnosis"[tiab] OR “diagnostic”) AND ((“clinical prediction"[tiab] OR “clinical model"[tiab] OR “clinical score"[tiab] OR “clinical scoring"[tiab] OR “validation of a clinical"[tiab] OR “decision guideline"[tiab] OR “validation study"[tiab] OR “validation studies"[tiab] OR “derivation study"[tiab] OR “screening score"[tiab] OR “decision rule"[tiab] OR “diagnostic rule"[tiab] OR “diagnostic score"[tiab] OR “predictive outcome"[tiab] OR “predictive rule"[tiab] OR “predictive score"[tiab] OR “predictive value"[tiab] OR “predictive risk"[tiab] OR “prediction outcome"[tiab] OR “prediction rule"[tiab] OR “prediction score"[tiab] OR “scoring"[tiab] OR “prediction value"[tiab] OR “prediction risk"[tiab] OR “risk assessment"[tiab] OR “risk score"[tiab] OR “risk scoring"[tiab] OR “prognostic score"[tiab] OR “prognostic index"[tiab] OR “prognostic rule"[tiab] OR “prospective validation"[tiab] OR (“risk"[tiab] AND “tool"[tiab]) OR ((“validate"[tiab] OR “validation"[tiab] OR “validating"[tiab] OR “develop"[tiab] OR “development"[tiab] OR “derivation"[tiab] OR “derive"[tiab] OR “deriving"[tiab] OR “performance"[tiab]) AND (“decision"[tiab] OR “predictive"[tiab] OR “prediction"[tiab] OR “rule"[tiab] OR “score"[tiab] OR “scoring"[tiab] OR “index"[tiab] OR “model"[tiab] OR “scale"[tiab] OR “tool"[tiab] OR “algorithm"[tiab])) OR (“development"[tiab] AND “validation"[tiab]) OR (“derivation"[tiab] AND “validation"[tiab]) OR “signs and symptoms"[tiab]))
NOT “Vaccine"
This search identified 4 potential studies not identified elsewhere for full text review.
A bridge search in June, 2021 identified 37 additional abstracts but none met our inclusion criteria.
Appendix 3. Definitions used for QUADAS-2 framework for quality assessment
Patient Selection
1. Was a consecutive or random sample of patients enrolled?
Y: Study enrolled consecutive patients or a random sample of consecutive patients with influenza symptoms from a primary care, urgent care, hospital, or ED setting.
N: A convenience sample or other nonconsecutive or nonrandom sample was used, or it only included patients referred for diagnostic imaging or to an ENT clinic (this does not address exclusion criteria, handled in #3 below).
U: Uncertain
2. Was the study designed to avoid a case-control design (Y/N/U)?
Y: The study population was drawn from a cohort that included patients with a spectrum of disease.
N: The study population consisted of patients with known disease and healthy controls
U: Uncertain
3. Did the study design avoid inappropriate exclusion criteria (Y/N/U)?
Y: There were no inappropriate exclusion criteria, such as excluding those with uncertain findings.
N: The study used inappropriate exclusion criteria.
U: Uncertain
4. Patient Selection Risk of Bias: What is likelihood that patient selection could have introduced bias (L/H/U)?
L: Low likelihood of bias due to patient selection or enrollment (“Yes” to 1, 2 and 3)
H: High likelihood of bias due to patient selection (“No” to 1, 2 or 3)
U: Unable to judge degree of bias.
5. Concerns About Patient Selection Applicability: Are there concerns that included patients and setting do not match the review question?
N: Patients are likely to be typical of patients with influenza symptoms.
Y: Patients are unlikely to represent typical patients with influenza symptoms, for example come from a specialized population, are immunosuppressed, etc.
U: Uncertain
Index Test: Influenza Symptoms
6. Were index test results interpreted without knowledge of reference standard?
Y: Yes (including when sign or symptom performed by clinician and reference standard performed using reference laboratory PCR that could not be known by clinician)
N: No
U: Uncertain
7. If a threshold was used for the index test, was it pre-specified?
Y: The threshold was prespecified or there was no threshold mentioned
N: The threshold was established post hoc
U: A threshold was used but it is not clear when it was specified
8. Index Test Risk of Bias: What is the likelihood that conduct of the index test could have introduced bias (L, H, U)?
L: Low likelihood of bias due to (“Yes” to 6 and “Yes” or “Uncertain” to 7)
H: High likelihood of bias due to failure to mask to reference standard (“No” to 6 or 7)
U: Uncertain
9. Concerns Regarding Index Test Applicability: Are there concerns that the index test differs from those specified in the review question?
L: Low likelihood (the index test is a commonly reported sign or symptom)
H: High likelihood (the index test is not a sign or symptom from history and physical examination, or comorbidity)
U: Uncertain
Reference Standard Test
10. Is the reference standard likely to correctly classify patients as having Influenza?
Y: Yes, used PCR.
N: No, used another reference standard
U: Uncertain
11. Was the reference standard interpreted without knowledge of the index test?
Y: Yes, reference standard interpretation masked to index test results
N: No, reference standard interpretation not masked to index test results
U: Uncertain
12. Reference Standard Risk of Bias: Could conduct or interpretation of the reference standard could have introduced bias?
L: Low likelihood of bias due to the reference standard (“Yes” to 9, “Yes” or “Uncertain” to 10)
H: High likelihood of bias due to inadequate reference standard (“No” to 9 or 10)
U: Uncertain
13. Concerns Regarding Applicability of the Reference Standard: are there concerns that the target conditions defined by the reference standard do not match the review question?
L: Low likelihood of bias, that is, the reference standard was intended to detect Influenza
H: High likelihood of bias, that is, the reference standard was not intended to detect Influenza
U: Uncertain
Patient Flow and Timing
14. Did all patients receive a reference standard?
Y: Yes, all patients received some sort of reference standard (no partial verification bias)
N: No, some patients did not receive any reference standard (partial verification bias)
U: Uncertain
15. Did all patients receive the same reference standard?
Y: Yes, all used the same reference standard (no differential verification bias)
N: No, the reference standard varied depending on the results of the index test (differential verification bias)
U: Uncertain
16. Were all patients included in the analysis?
Y: All patients were properly accounted for in the analysis
N: Some patients were not accounted for or dropped for unclear reasons
U: Uncertain
17. Patient Flow Risk of Bias: Could patient flow have introduced bias?
L: Low likelihood of bias based on absence of partial verification bias and good follow-up (“Y” on 14 and15, “Yes” or “Uncertain” for 16)
H: High likelihood of bias based on partial verification bias or poor follow-up (“No” to 14 or 15, or significant number of patients lost to follow-up in 16)
U: Uncertain
Appendix 4. Clinical Prediction Rules Included in the Systematic Review
Notes
This article was externally peer reviewed.
Funding: The Seattle Flu Study is funded by Gates Ventures. The funder was not involved in the design of the study, does not have any ownership over the management and conduct of the study, the data, or the rights to publish.
Conflict of interests: None.
To see this article online, please go to: http://jabfm.org/content/34/6/1123.full.
- Received for publication March 15, 2021.
- Revision received June 8, 2021.
- Accepted for publication August 19, 2021.