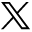Abstract
Introduction: The goal of public health infectious disease surveillance systems is to provide accurate laboratory results in near-real time. When it comes to influenza surveillance, most current systems are encumbered with inherent delays encountered in the real-life chaos of medical practice. To combat this, we implemented and tested near-real-time surveillance using a rapid influenza detection test (RIDT) coupled with immediate, wireless transmission of results to public health entities.
Methods: A network of 19 primary care clinics across Wisconsin were recruited, including 4 sites already involved in ongoing influenza surveillance and 15 sites that were new to surveillance activities. Each site was provided with a Quidel Sofia Influenza A+B RIDT analyzer attached to a wireless router. Influenza test results, along with patient age, were transmitted immediately to a cloud-based server, automatically compiled, and forwarded to the surveillance team daily. Weekly counts of positive influenza A and B cases were compared with positive polymerase chain reaction (PCR) detections from an independent surveillance system within the state.
Results: Following Institutional Review Board (IRB) and institutional approvals, we recruited 19 surveillance sites, installed equipment, and trained staff within 4 months. Of the 1119 cases tested between September 15, 2013 and June 28, 2014, 316 were positive for influenza. The system provided early detection of the influenza outbreak in Wisconsin. The influenza peak between January 12 and 25, 2014, as well as the epidemic curve, closely matched that derived from the established PCR laboratory network (r = 0.927; P < .001).
Conclusions: A network of influenza RIDTs with wireless transmission of results approximated the long-sought-after goal of real-time influenza surveillance. Results from the initial year strongly support this approach to highly accurate and timely influenza surveillance.
- Communicable Diseases
- Disease Outbreaks
- Influenza
- Polymerase Chain Reaction
- Practice-based Research
- Primary Health Care
- Public Health Surveillance
- Wisconsin
The goal of public health infectious disease surveillance is to provide meaningful information on circulating pathogens in a timeframe that facilitates intervention.1,2 Along with the usual demographic, clinical and epidemiologic descriptors, components of meaningful information include the sensitivity, specificity, and accuracy of pathogen detection. Additional attributes of equal importance for surveillance systems include timeliness, flexibility, and cost. Unfortunately, the quality of results and the timeliness, flexibility, and cost of surveillance systems are often diametrically opposed, thus, necessitating tradeoffs.
Current approaches to influenza surveillance are based on sentinel clinician networks, laboratory detection and reporting, and mechanistic tools.3⇓–5 Sentinel networks engage trained clinicians to identify influenza-like illness using prespecified clinical criteria.6⇓⇓⇓–10 Such efforts can be sensitive and flexible, but may also be costly and suffer from low specificity.11 Use of influenza laboratory results is highly specific for influenza and can be flexible, but is costly and relatively insensitive, given that testing is usually limited to individuals meeting clinical criteria.12,13 Mechanistic surveillance—in which existing data are aggregated and analyzed for surveillance purposes, such as the pneumonia and influenza mortality index and Google Flu Trends—is inexpensive, flexible, and sensitive, but can be limited by a lack of specificity.14⇓–16
For most systems, timeliness has been a major limitation due to inherent delays in manual reporting, data aggregation, and result dissemination. Participatory mechanistic surveillance, which captures voluntarily submitted symptom data from the general public, has been shown to be timely, but is limited in specificity.14,15 Likewise, Internet surveillance based on usage patterns (eg, Google, Wikipedia, Twitter) could improve timeliness for detecting health events such as influenza, but the approach is still relatively new and has significant limitations.16⇓–18
For this report, we focused on an innovative, novel approach to influenza surveillance using sensitive and specific point-of-care testing in primary care settings19,20 coupled with anonymous results sent immediately to a server through wireless technology. Daily results were aggregated and forwarded to the surveillance team and to public health personnel, thus, providing a solution to common reasons for reporting delays and allowing for near-real-time surveillance.
Methods
Timeframe
The concept for this pilot and feasibility study emerged from an initial meeting of the surveillance team on May 2, 2013. The goal was to develop a surveillance program to be in place for the 2013 to 2014 influenza season. Data collection began September 15, 2013 and concluded June 28, 2014.
Setting
The base of this network included 4 existing influenza surveillance clinics located in south-central Wisconsin that are affiliated with the University of Wisconsin, Department of Family Medicine and Community Health. The clinics had been enrolled in the Influenza Incidence Surveillance Project (IISP), an outpatient system funded by the U.S. Centers for Disease Control and Prevention (CDC),21,22 for 3 to 5 years. We engaged the Wisconsin Research and Education Network (WREN),23 a long-standing practice-based research network, to identify and recruit 16 additional primary care practices across Wisconsin. The goal was to recruit at least 2 sites in each of the state's 5 public health regions. Practice ownership, number of clinicians, existing use of rapid influenza detection tests (RIDT), or use of electronic health records were not considered in site selection.
Participants
The participants included family physicians, pediatricians, physician assistants, and nurse practitioners. More than 250 clinicians and clinical staff received brief training regarding the selection of patients, nasal specimen collection, and interpretations of RIDT results. Our intent was to deliberatively introduce a high level of variability and chaos into the overall pilot to replicate real-life medical practice.24
Clinician Surveillance Protocol
Two parallel surveillance protocols were used, but the differences primarily involved the types of specimens collected. At all sites, clinicians were asked to identify patients of any age who presented with acute respiratory infections (ARIs) regardless of the reason for visit (eg, well care, followup, acute care). ARIs were characterized by at least 2 of the following symptoms: fever, cough, sore throat, nasal congestion or runny nose, and with onset of symptoms starting no more than 4 days before visit. It was up to individual clinicians to define fever and determine whether, on clinical grounds, it was appropriate to obtain a specimen using a nasal foam swab. Clinicians were instructed to identify the nostril with the most secretion (if present), insert the foam swab approximately 1 inch, gently rotate the swab a few times, place swab back into sheath, affix a patient label onto the sheath, and send it to the onsite clinical laboratory for immediate processing via Quidel Sofia Influenza A+B RIDT. Results were confirmed through polymerase chain reaction (PCR) testing at the Wisconsin State Laboratory of Hygiene (WSLH).
For the 4 pre-existing IISP sites, an additional oropharyngeal or nasopharyngeal specimen was obtained from each patient using an appropriate swab and placed into viral transport media (VTM) for influenza PCR testing at the WSLH. Due to existing IISP protocols, these 4 sites allowed specimen collection up to 7 days after symptom onset. In addition, patient demographic information, epidemiologic data, symptoms, and vaccine history were obtained at these sites as part of the existing Influenza Incidence Surveillance Program (IISP) protocol.21,22
Laboratory Procedures
WREN staff members trained the clinical laboratorians at each WREN site to use Quidel Sofia Influenza A+B RIDT.25 All laboratory procedures followed the package insert.26 Following the usual specimen processing, the used nasal or “residual” swab was placed into a labeled VTM tube at the WREN sites. A WSLH requisition form was completed, which included the results from Sofia Influenza A+B testing. The VTM tube was then placed with a cool pack to maintain temperature at 4° to 8°C, and was shipped with the requisition form to WSLH by courier.
Rapid Influenza Detection Test
We used the Quidel Sofia Influenza A+B Fluorescent Immunoassay RIDT.25,26 Sofia uses advanced immunofluorescence-based lateral-flow technology to detect influenza A and B viral nucleoprotein antigens in nasal specimens. The analyzer scans the test strip, measures the immunofluorescent signal, and displays the test results (Positive, Negative, or Invalid) on its display screen. This low-complexity RIDT is Clinical Laboratory Improvement Amendments (CLIA) waived for nasal and nasopharyngeal swabs and provides results in 20 minutes. Nasal swab sensitivity and specificity per the package insert are 90% and 95% for influenza A and 89% and 96% for influenza B, respectively.26
Data Flow
The Sofia analyzers were equipped with wireless transmitters, using the Verizon network, to transmit anonymous test result data to Virena.27 Virena is a propriety, global wireless surveillance and remote instrument data management system that provides a near-real-time deidentified database. Transmitted data included location (device identifier), date and time of testing, patient age, and results of influenza A and B testing. All data were automatically compiled in an Excel spreadsheet and emailed to the surveillance team daily.
Data Analyses
Simple graphical tools were developed in Excel to allow cutting and pasting of daily data files from Virena into an analytic spreadsheet. Automated processes, developed using Excel's macro feature, allowed numeric and graphical assessments of influenza detection by clinic, public health region, and statewide. We assessed the percent positivity of specimens by week, and applied 3-point and 5-point moving averages to smooth curves. Data and results were summarized on a weekly basis and weekly feedback reports, which included a graph of influenza A and B results, were provided to all participants.
Comparisons were made to a longstanding statewide influenza surveillance system coordinated by the WSLH in collaboration with the Wisconsin Division of Public Health.28 The WSLH PCR Laboratory Network compiles influenza PCR results from clinical laboratories across Wisconsin as well as surveillance specimens tested at WSLH. We compared the weekly counts of positive results from our network with those of the WSLH-PCR network using Pearson correlation.
Human Subjects Protections
This protocol was found to be exempt from Institutional Review Board (IRB) oversight by the University of Wisconsin Health Sciences Institutional Review Board. In addition, the protocol was exempted by 7 additional IRBs associated with WREN clinics. These findings were based on a determination of usual clinical care using an U.S. Food and Drug Administration (FDA) approved test coupled with public health surveillance.
Results
Creation of Statewide Network
Identifying, recruiting, equipping, and training for the overall program were accomplished in a relatively short timeframe. Following the initial concept meeting in early May 2013 and University of Wisconsin Health Sciences IRB exemption on June 25, 2013, recruitment was stalled until a funding contract was approved by the UW Board of Regents on August 20, 2013. At that point WREN recruited participants rapidly. Progress was then delayed by secondary IRB reviews at 7 sites. Despite delays imposed by external review, 19 of 20 anticipated sites were up and running by December 2013.
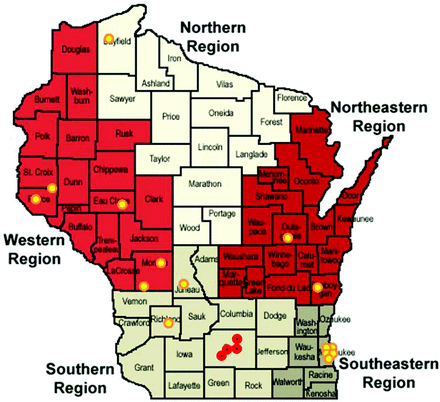
At least 1 clinic was recruited from each of the 5 public health regions in Wisconsin (Figure 1). The clinics varied in size, personnel, and ownership (Table 1). At least 258 clinic staff (range, 3 to 38 per site) were trained on the study protocol and included clinicians (MD, DO, PA, NP, CNM), nurses, medical assistants, laboratory staff, pharmacists, dieticians, and administrative staff (reception, clinic managers).
Location of Real-Time Influenza Surveillance Network sites. WREN practices are indicated by yellow dots. IISP clinics are shown by red dots.
Characteristics of Participating Clinics in the Real-Time Influenza Surveillance Network
There were some initial complications, including lack of wireless service at 1 site, difficulty with transmission from a site where the laboratory was in a basement, 3 nonfunctional wireless units that needed replacement, and identifying proficiency testing results among clinical results.
Surveillance Population
Although all clinics performed specimen collection and testing, roughly half of the specimens originated from the 4 IISP clinics and the number of specimens per site varied from 4 to 205 with a median of 43. Nasal specimens were obtained from 1133 patients, age 1 month to 93 years. Nine specimens were not tested by Sofia and 5 had invalid Sofia results (0.4%), yielding a final surveillance population of 1119 cases. The mean age (± SD) was 32.3 ± 20.9 years with a median of 33.1 years. A composite view of the population demonstrates peaks of ARI patients in early childhood and in the late 20s and early 30s (Figure 2).
Results by age of surveillance population, showing cases with negative Sofia RITD results and those with influenza A and influenza B.
Detection of Influenza
There were 251 influenza A positives (22.2%) and 62 influenza B positives (5.5%). Three specimens were positive for both influenza A and B (0.3%). The average age of influenza A–positive patients was 29.6 ± 20.5 years (median, 31 years; range, 2 months to 82 years) and the average age of influenza B–positive patients was 33.8 ± 22.5 years (median, 35 years; range, 2 to 78 years).
A dramatic increase in influenza A–positive specimens occurred December 8 to 14, after which detections continued to increase until a peak was reached January 12 to 25. Influenza A declined to baseline March 9 to 15. The epidemic curve is depicted in Figure 3.
Epidemic curve for influenza cases as detected by the Real-Time Influenza Surveillance Network beginning September 15, 2013 and ending June 28, 2014.
Comparison with Existing Surveillance
Excellent comparability was noted between the real-time network and the existing surveillance system in Wisconsin, despite a much reduced sample size in the former (Figure 4). A highly significant correlation was found between the weekly real-time network detections of influenza A and that of the WSLH-PCR network (r = 0.927; P < .001; Figure 5).
Epidemic curves for influenza A. Data are expressed as the percent of the maximum weekly influenza case count for each system. RTISN counts (n = 254) were available immediately; WSLH-PCR counts (n = 4780) were delayed by 1 to 2 weeks.
Correlation of weekly influenza A case counts between Real-Time Influenza Surveillance Network and the WSLH PCR Network.
Discussion
Combining rapid, sensitive, and specific point-of-care testing with wireless technology in primary care provides real-time surveillance of influenza. We demonstrated that the insertion of this novel technology into ambulatory care practices can produce a high-quality and timely representation of influenza across a state despite minimal training and high levels of variation and chaos that is common in primary care.24 Moreover, surveillance covered a broad and representative population and reflected the expected community trends in ARI.29 The system provided extremely early detection for the 2013 to 2014 seasonal influenza outbreak in Wisconsin. The resulting epidemic curve had excellent concordance with the curve based on the existing statewide laboratory surveillance system, with close matching of the onset, peak, and conclusion of the influenza outbreak. Of note is the 1-to-2-week time delay required to aggregate data and to generate the comparator surveillance data from the PCR network by the WSLH.
We also demonstrated that this technology could be inserted rapidly into busy primary care practices, most of which were not using any RIDTs. Despite the time encumbrances imposed by a new research protocol, contract negotiations, 8 separate IRB reviews, recruiting, equipment deployment, and training, we achieved a functional surveillance network in very little time. This was facilitated using WREN practices and underscores the value of established relationships within practice-based research networks.30,31 A similar surveillance program could, therefore, be easily implemented as a public health response system using clinics already familiar with RIDTs, thus omitting all the steps noted above.
The 2013 to 2014 influenza season was dominated by influenza A (H1N1). Across the United States, influenza A viruses accounted for 87.4% of all influenza detections reported by the CDC compared with 12.6% for influenza B.32 Our Sofia data were similar with influenza A and B representing 80.2% and 19.8% of positive detections, respectively.
This feasibility study had some important limitations. First, we noted wide variation in clinic participation, which was likely due to intentionally limited oversight by project coordinators and IISP clinics already being accustomed to the process. Such locational bias can affect features of the epidemic curve. For example, early activity in 2013 occurred in southeast Wisconsin, approximately 2 weeks before the onset in south-central Wisconsin. Second, there was no attempt to define the clinical population denominator necessary to evaluate incidence and prevalence. Ongoing assessments of influenza were based on percent positive rates of tests (positive results divided by all tests performed) and weekly counts of influenza cases. Third, our results are based on Sofia. The performance of other specific RIDTs may alter surveillance results. Accordingly, confirmatory testing of some specimens is recommended. Fourth, the diagnostic sensitivity of the RIDT in actual use tends to be considerably lower than values listed on the product insert, resulting in false negatives.25,26 Lower sensitivities, however, are tolerated in surveillance systems due to the ability to aggregate data. Finally, unanticipated technological problems were encountered, which occasionally prohibited wireless transmission.
Many of the limitations, however, had positive attributes. The study was designed to be a pragmatic trial in which great variation and chaos—common attributes of real-life primary care—were courted and embraced. The surveillance results were excellent despite this background “noise.” Collection of residual nasal swab specimens in VTM allowed confirmatory testing with RT-PCR and direct evaluation of sensitivity and specificity. Discussion of performance characteristics of Sofia, however, is beyond the scope of this article.25,33,34
Although not explored systematically in this study, clinical sites may have received some added benefits from participation. As part of our usual operation, we provided weekly feedback on the circulation of influenza and other respiratory viruses in Wisconsin. Although we did not collect clinical responses to rapid influenza detection, previous studies have shown information on respiratory viruses can assist in diagnosis and reduce antibiotic prescribing.35 Electronic transfer of syndromic surveillance data to public health agencies is a metric used in attaining patient-centered medical home certification.36 Participation in a surveillance network, such as we piloted in this study, could be used to meet this requirement.
In this study, the Sofia analyzers, wireless arrays, and test kits were provided by the manufacturer. Although we did not perform any formal economic assessment, the cost of such a network, especially if built with practices already using RIDTs in usual practice, is likely not prohibitive. The additional cost to a public health department could include the wireless transmitters and the monthly wireless bill. In our situation, the cellular costs were approximately $1,400 per month for the entire network. Discussion on the number of such sites per geographic area and the appropriate geospatial deployment are beyond this discussion, but have been evaluated by the Association of Public Health Laboratories as part of the Influenza Virologic Surveillance Right Size Roadmap.37
Although it is still necessary to confirm results through PCR testing to meet public health accuracy standards, Sofia RIDT coupled with wireless transmission of anonymous test results creates real-time surveillance of influenza with the free flow of result data to public health agencies in near real time. Automated processes allow for data to be aggregated daily, electronically pushed out, and analyzed on a daily and weekly basis. Such a system eliminates the need for anyone to assemble, aggregate, and send information and, thus minimizes the lag times inherent in reporting. It can be implemented quickly in primary care settings, especially when working with a practice-based research network. As a public health activity, IRB approval would generally not be needed as this system couples routine clinical care with anonymous reporting to public health agencies. Extremely simple surveillance protocols allow buy-in from clinics and clinicians. This approach also creates robust and reliable surveillance information that can achieve extremely early outbreak detection. Finally, this is a highly adaptable system that could incorporate other target pathogens for which RIDT technology is, or becomes, available.
Acknowledgments
We thank the practices and clinicians of the Wisconsin Research and Education Network (WREN) for participation, and staff members, Melody Bockenfeld, Erin Legee, Kate Judge, and Amy Irwin for outreach and site support. Mary Wedig and laboratory staff of the Wisconsin State Laboratory of Hygiene provided data management and excellence in laboratory testing and proficiency testing. Tom Haupt of the Wisconsin Division of Public Health assisted in project oversight. Cristalyne Bell contributed to manuscript revision. Finally, David Booker of Quidel Corporation provided invaluable technical assistance with wireless connectivity and troubleshooting.
Notes
This article was externally peer reviewed.
Funding: This study was funded through a research grant from Quidel Corporation to the Board of Regents at the University of Wisconsin.
Conflict of interest: JT is employed by Quidel Corporation. JLT received research funding from Quidel Corporation. The study idea emerged from a meeting between JLT, PS, and JT on May 2, 2013. Quidel Corporation did not direct or exert any influence over this manuscript.
To see this article online, please go to: http://jabfm.org/content/30/5/615.full.
- Received for publication February 7, 2017.
- Revision received June 2, 2017.
- Accepted for publication June 8, 2017.